Introduction to LinkML
Whilst trying to define an open-source data model for NHS pay, I was initally stumped on which technology to use. I wanted to build something that would support downstream developers, regardless of their preferred tech stacks, trying to build a ‘general purpose’, abstract data model.
As I was weighing up the pros and cons of things like JSON-schema, Pydantic and OWL (among others), I kept lamenting the following: given all of these data formats are conceptually interoperable, why on earth hadn’t someone built a good modelling language that could serialize into all the other formats?
Turns out, someone had! Enter, LinkML - exactly what I’d been looking for.
LinkML features
LinkML describes itself as a “a general purpose modeling language that can be used with linked data, JSON, and other formalisms”. The thing that’s really cool about it, is that it provides generators for compiling this data model to other frameworks.
I will not walk through how it works, or how to use it. I’ll instead leave that to the excellent documentation. What I will do, however, is highlight the core features I love about it.
Model once, validate everywhere
One of the first exercises in the documentation walks you through creation of a simple data model, and allows you to validate arbitrary data formats. You can write a model in YAML, and validate an object instance that’s defined in JSON, with ease.
Easy transformations
In the same step (within ~1 minute’s worth of reading) you’ve already got to the next feature, where you can translate a object in one format (e.g. JSON) into another (e.g. RDF Turtle), with a single command. Cue a small tear being brought to my eye.
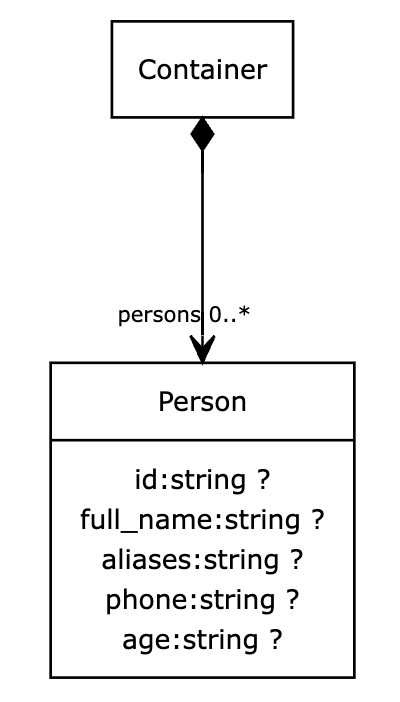
Visualization
Data visualization is, to me, a very important part of data modelling. As well as making it easier to communicate with domain experts that may not be comfortable reading markup, I also find it a good cognitive aid.
LinkML allows you to easily create UML diagrams from a data model, such as the one below:
Semantic markup
The final addition that one me over for LinkML, is that it provides you with the ability to back up your schema with semantic annotations. As mentioned here, all the elements of a LinkML schema (i.e. class and field definitions) have a globally unique IRI/URI. This is either explicitly defined by the authoring individual, or implicity, using the default prefix.
The benefit of doing this may not be familiar to all readers, but it is core tenant of the semantic web community. URIs are just a way to uniquely label an arbitrary (or ‘resource’) which, among other things, is helpful for performing entity resolution. This article talks more about it for those that are interested.. It can be a bit meta, and not strictly necessary for many use cases, but it’s great to have the option to expand the model with such annotations if needed.
Conclusion
Once again, it seems the biology community is crushing it with respect to linked data tooling. I was unreasonably excited to discover LinkML, and imagine I will be using it in many projects down the line.